Configuration Options¶
Setup Galaxy Server for Blast processing.
Configuration Files¶
globals.js¶
Modify the configuration file as necessary.
To view aggregate configuration: ./jbutil --config
The aggregate config file is the merged config of JBConnect and its installed jbconnect-hook-* modules.
Edit config file: nano config/globals.js
module.exports.globals = {
jbrowse: {
galaxy: {
galaxyUrl: "http://localhost:8080", // URL of Galaxy
galaxyPath: "/var/www/html/galaxy", // path of Galaxy
//galaxyPath: "/var/www/html/galaxy_jblast", // path of Galaxy, if docker
galaxyAPIKey: "c7be32db9329841598b1a5705655f633", //
// jblast will use this Galaxy history
historyName: "Unnamed history" // default history name
},
jblast: {
blastResultPath: "jblastdata",
blastResultCategory: "JBlast Results",
insertTrackTemplate: "inMemTemplate.json",
import: ["blastxml"]
}
}
};
blastResultPathis the sub directory within the dataset directory where the blast results are storedblastResultCategoryis the name of the JBrowse track selectory category.insertTrackTemplateis the track insertion template.importis the file extension to process from the Galaxy workflow.
Standalone Blast Job Service¶
The job service, basicWorkdlowService, is the job runner service that manages stand-alone Blast processing.
Requirements¶
These requirements are generally installed as part of the JBlast project.
npm install enuggetry/blast-ncbi-tools (NCBI blast)
npm install enuggetry/faux-blastdb (a small sample blast database)
Configuration¶
Add the following to the jbconnect.config.js file, enabling basicWorkflowService:
module.exports = {
jbrowse: {
services: {
'basicWorkflowService': {enable: true, name: 'basicWorkflowService', type: 'workflow', alias: "jblast"},
'galaxyService': {enable: false, name: 'galaxyService', type: 'workflow', alias: "jblast"}
},
}
};
featureMapping flag¶
featureMapping defines how the target features mapped into coordinate space.
‘hit’ - target features will be mapped into target coordinates. ‘query’ (default) - target features will be mapped into query space on the in the contig where the query is made.
If featureMapping=’hit’, then contigHandler() function will be needed to decipher the proper contig of the feature. If featureMapping=’query’, then the contigHandler() is ignored.
dataSet: {
ChineseSpring: { path: 'IWGSC',
featureMapping: 'hit',
contigHandler(hit) {
console.log("IWGSC contigHandler");
return hit.Hit_accession;
}
},
},
The contigHandler() function allows for selecting the value from a hit will be treated as the sequence id of the feature. The hit is passed to the function.
Blast Profiles¶
Blast profiles are parameter lists that translate to NCBI blast command line parameters sets.
For example: for the following blast command
blastn –db nt –query nt.fsa –out results.out
the blast profile would look like:
'myprofile': {
'db': 'nt',
'query': 'nt.fsa',
'out': 'result.out'
},
config/globals.js defines the blast profiles that are preconfigured with jblast.
(note, only faux blast database is autoatically loaded by JBlast project. So, ‘htgs’ shouldn’t be used unless htgs blast database is first installed.)
jblast:
defaultBlastProfile: 'faux',
blastProfiles: {
'htgs': {
'db': 'htgs'
},
'faux': {
'db': 'faux'
},
'remote_htgs': {
'db': 'htgs',
'remote': ""
}
}
}
The blastProfile can be specified the POST /job/submit. For example:
var postData = {
service: "jblast", // this can be the name of the job service or its alias
dataset: "sample_data/json/volvox",
region: ">ctgA ctgA:44705..47713 (- strand) class=remark length=3009\nacatccaatggcgaacataa...gcgagttt",
workflow: "NCBI.blast.workflow.js"
blastProfile: 'faux' // selects the 'faux' profile that is defined in globals.js.
};
$.post( "/job/submit", postData , function( result ) {
console.log( result );
}, "json");
Alternatively, an previously undefined profiled may be specified in /job/submit.
var postData = {
service: "jblast", // this can be the name of the job service or its alias
dataset: "sample_data/json/volvox",
region: ">ctgA ctgA:44705..47713 (- strand) class=remark length=3009\nacatccaatggcgaacataa...gcgagttt",
workflow: "NCBI.blast.workflow.js"
blastProfile: {
'db': 'nt',
'query': 'nt.fsa',
'out': 'result.out'
}
};
$.post( "/job/submit", postData , function( result ) {
console.log( result );
}, "json");
If defaultBlastProfile is defined in globals.js will be used if no blast profile is specified in the /job/submit call.
Blast profiles only apply to basicWorkflowService.
JBrowse Peer Configuration¶
JBrowse can be configured in a peer directory instead of a module. In this case, bypass the Install JBrowse and Setup Demo step.
For example:
/home
/zuser
/JBConnect
/jbrowse
Create a file called jbconnect.config.js in the JBConnect app directory that contains the following:
module.exports = {
jbrowse: {
jbrowsePath: "/home/zuser/jbrowse/"
}
};
Galaxy Blast Job Service¶
The galaxyService requres the presence of Galaxy.
See Setup Galaxy for instructions on how to configure Galaxy for JBlast.
JBlast jbutil Command¶
jbutil is a setup/configuration utility for JBConnect. JBConnect hooks can extend
jbutil command options. (see: jbs-hooks-extend)
This example shows that jbconnect-hook-jblast adds a number of commands to jbutil
$ ./jbutil --help
Usage: node jbutil
-c, --config display merged configuration
--setupworkflows (jblast-galaxy) installs demo galaxy workflows (must have API key configured
--setuptools (jblast-galaxy) setup jblast tools for galaxy
--setupdata (jblast) setup jblast demo data and samples
-o, --overwrite (jblast) used with --setupdata - overwrite samples
-d, --dbreset reset the database to default and clean kue db
-f, --force --dbreset without verifying
-a, --setadmin set admin flag
-r, --removeall remove JBConnect components from JBrowse
--pushplugins link plugins into JBrowse dir
--coverage=PLUGIN used with --pushplugins to add coverage instrumentation
--buildwebpack build jbrowse webpack
-h, --help display this help
–setupworkflows¶
This option setus up sample JBlast workflows in galaxy. This requires having configured the Galaxy API key in config.
–setuptools¶
This option sets up Jblast tools for Galaxy. After this is called, Galaxy will need to be restarted.
Note: NCBI Blast tools are not installed by the ``jbutils –setuptools`` script. the user must manually install these through the Tool Shed as admin.
–setupdata¶
This options sets up samples and sample data for JBlast.
JBlast Plugin¶
JBlast has integrated GUI features that must be enabled with by installing the JBlast plugin
and the JBClient on the client side.
In trackList.json, within the dataset’s path, add JBlast and JBClient plugin to the configuration.
"plugins": [
"JBClient", <-----
"JBlast", <-----
"NeatHTMLFeatures",
"NeatCanvasFeatures",
"HideTrackLabels"
],
Note: the JBlast and JBClient plugins are not physically in the JBrowse plugin directory. They are made available as route by the JBConnect framework and are only accessible at runtime.
See Integrated GUI for more details.
Limiting the BLAST query size¶
In dataset’s trackList.json, define bpSizeLimit to limit the size of the sequence query.
"plugins": [
...
"JBClient",
{
"name": "JBlast",
"bpSizeLimit": 15000
}
],
Sample Sequence Button¶
Configure a Sample Sequence Button in the BLAST a DNA sequence dialog box.
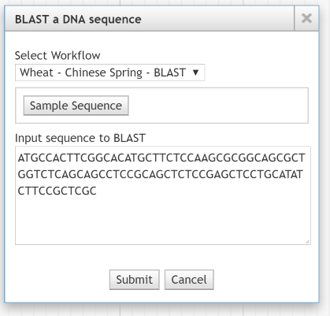
BLAST DNA Sequence Dialog box
Add the following section in the JBrowse trackList.json file.
"demo": {
"blastButtons": [
{
"button": "Sample Sequence",
"description": "Insert sample sequence",
"sequence": [
"ATGCCACTTCGGCACATGCTTCTCCAAGCGCGGCAGCGCTGGTCTCAGCAGCCTCCGCAGCTCTCCGAGCTCCTGCATATCTTCCGCTCGCTCTCTATTCTA",
"CCATCCGGAGCCTCTGCCGCGCCATCCAACCGCTCTCCTAGGCAAATTCAGCTGCCCCAAACCTTGCCTTGCTCCAATGCCAACCCACTTGGCGCCGGCTTC",
"CACATCGACGTTGTTGACGATGACCTCTGGCCCACTTCCTTCGGCTTCTCCTCAGATCCCATGACTGGTGATGAGTGTCTTGATACCTTCCAAGAACACGGA",
"GAAGAACAAGTGCACGACTCGGATGATGAGATAGATGACATGAGGCACCGCAAGCAGCTGTTCTACAAGCTGGACAGGGGGTCCAAGGAGTTTGAGGAATAT",
"AACTTGCCCTTGCGCCGCAGATGGAAGAGAGATAAACCCAATGCCAAGAATCCATCCGATTGCG"
]
}
]
},